
|
|
|
Status:
Completed
Views
324
Topics
Referenced by
Cite this as
Daniel Himmelstein, Lars Juhl Jensen (2015) One network to rule them all. Thinklab. doi:10.15363/thinklab.d102
License
Share
|

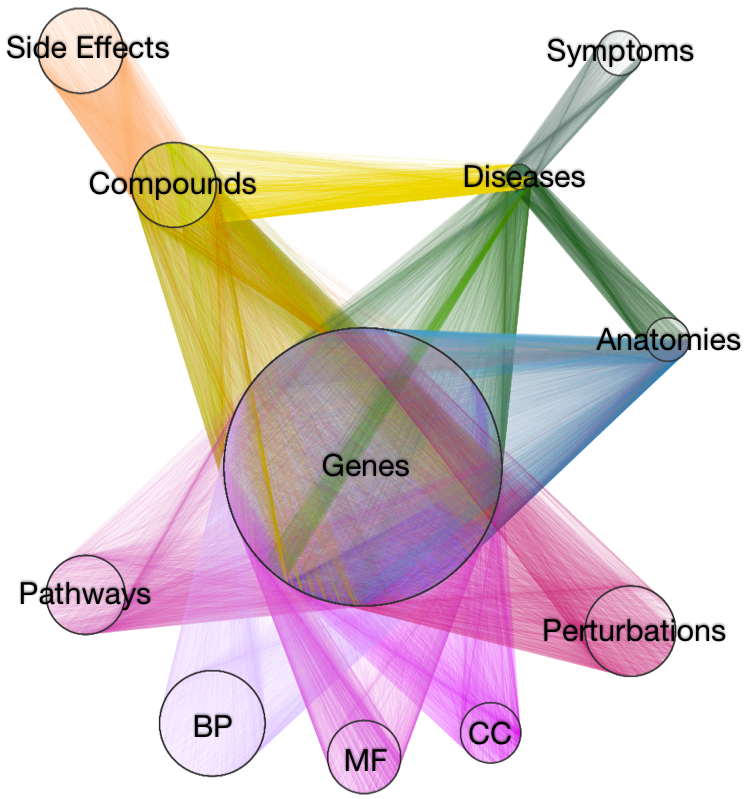

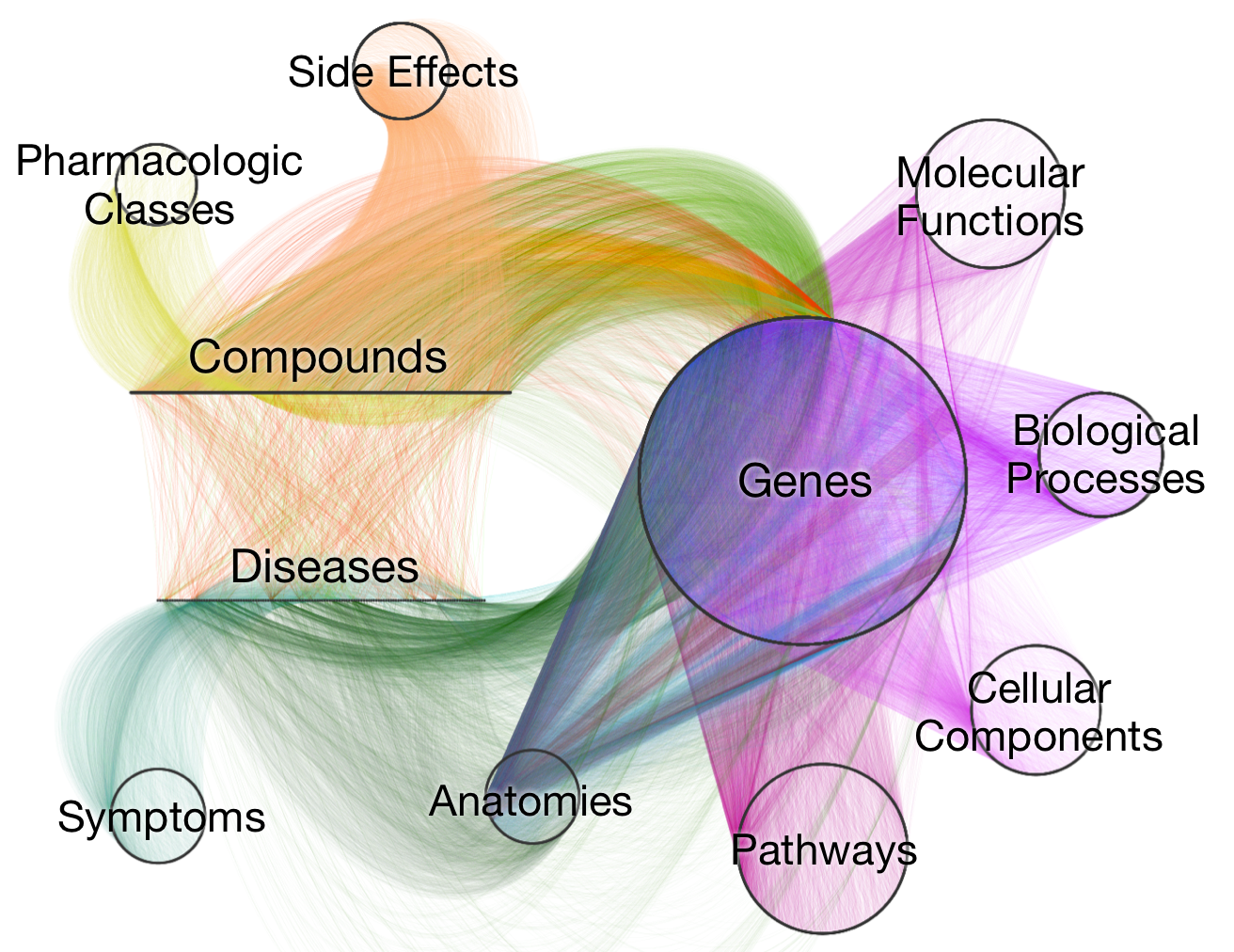

Daniel Himmelstein: 2015-09-06: I updated this post to correct edge counts. Previously, I wrote the network contains "5,998,711 edges (2,977,167 of which are unbiased)". The issue was caused by counting single edges multiple times.
Daniel Himmelstein: Here are two references for publishing hashes of academic content to the bitcoin blockchain: a 2014 blog post and a 2016 paper [1].