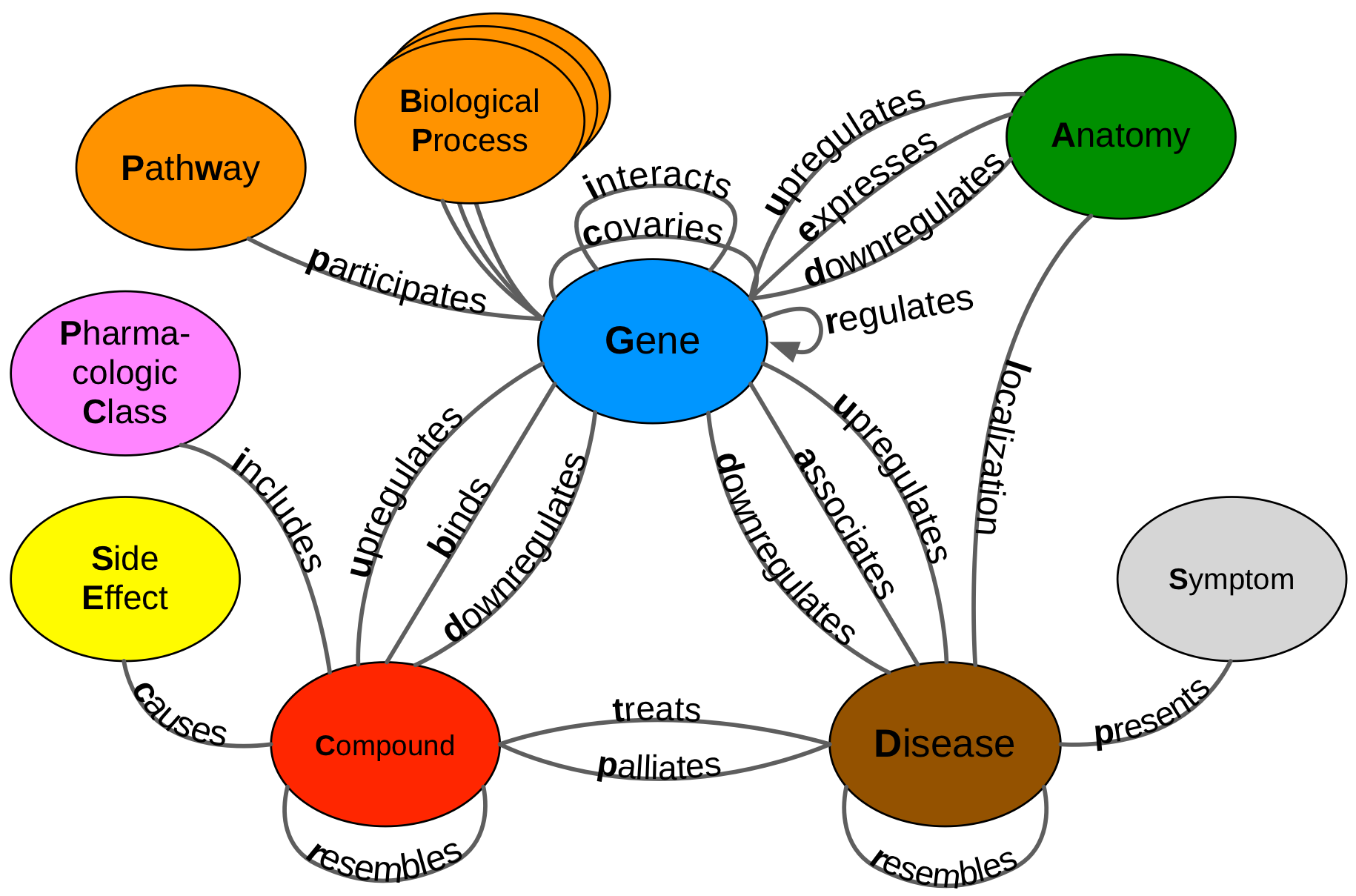
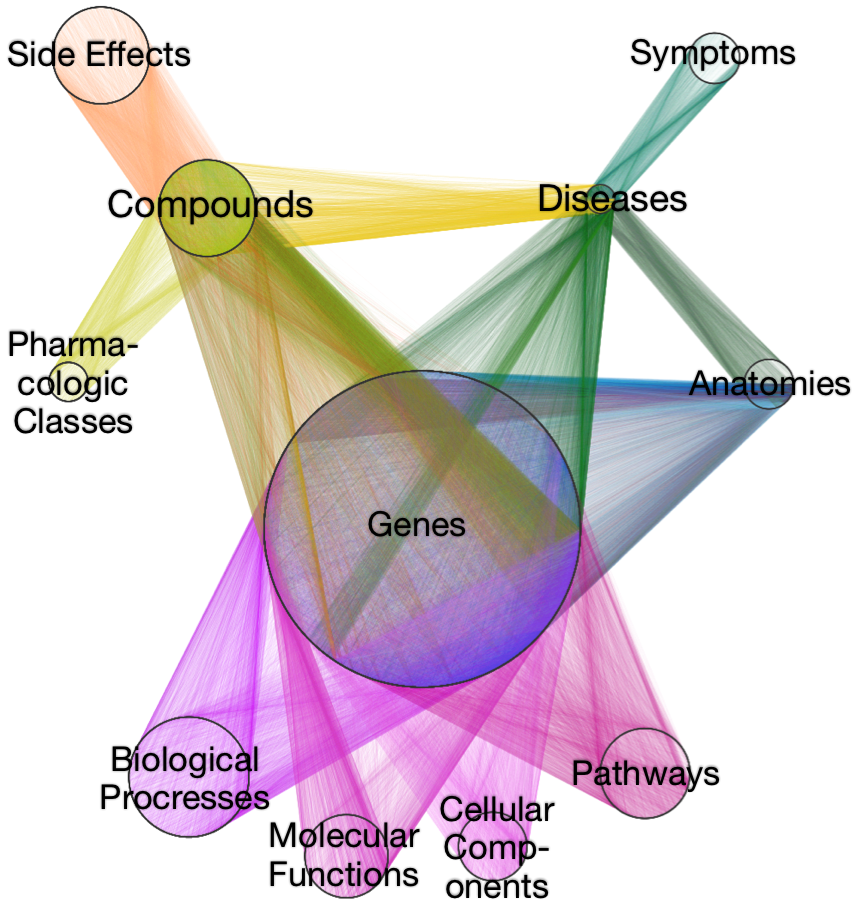
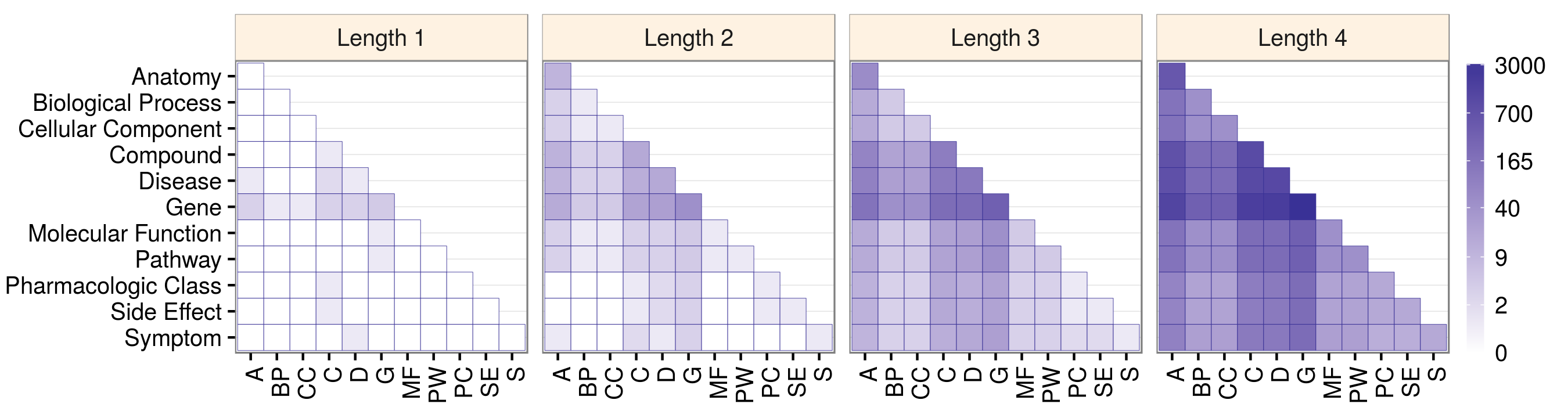
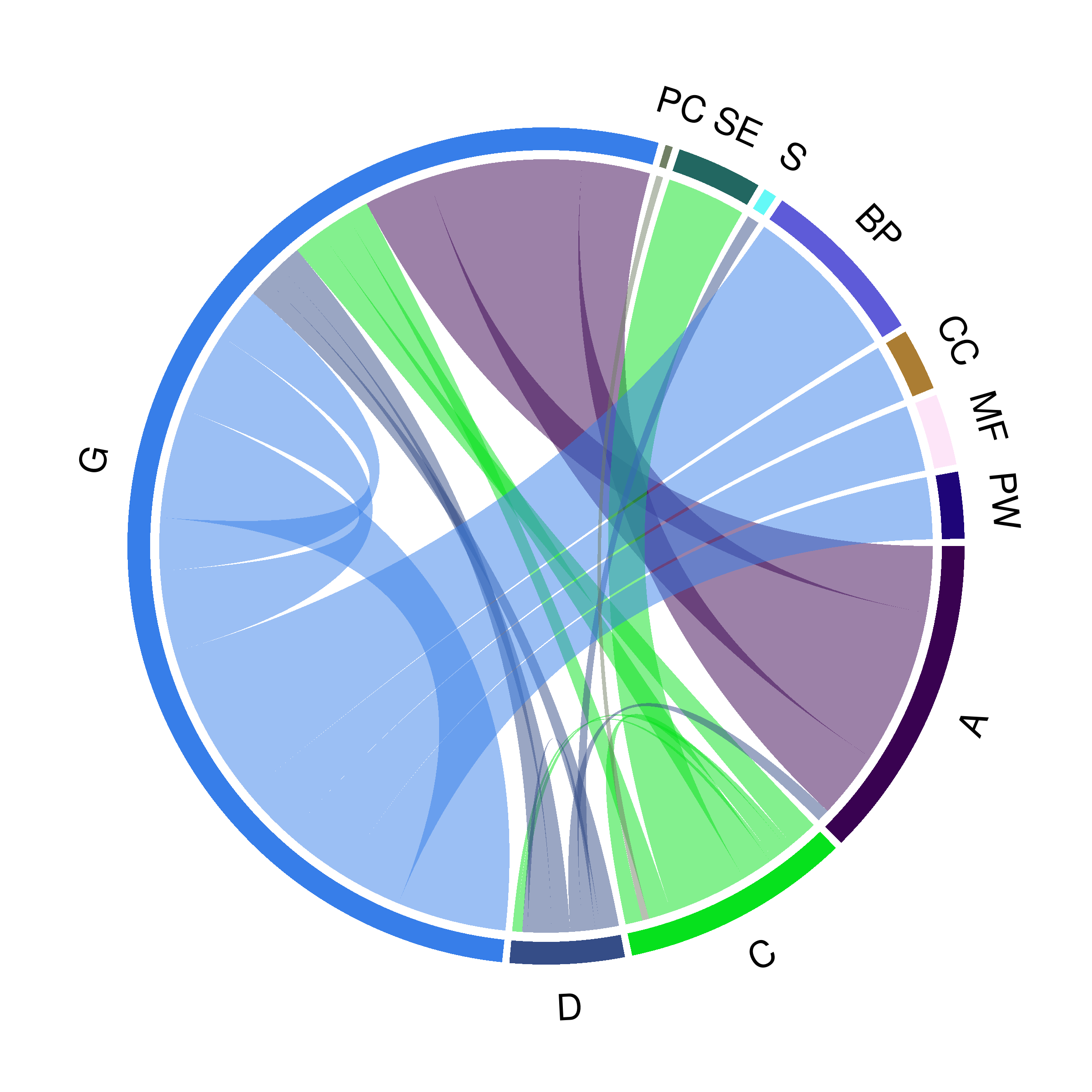
|
|
|
Status:
Open
Views
225
Topics
Referenced by
Cite this as
Daniel Himmelstein, Sergio Baranzini, Casey Greene (2016) Describing Hetionet v1.0 through visualization and statistics. Thinklab. doi:10.15363/thinklab.d202
License
Share
|