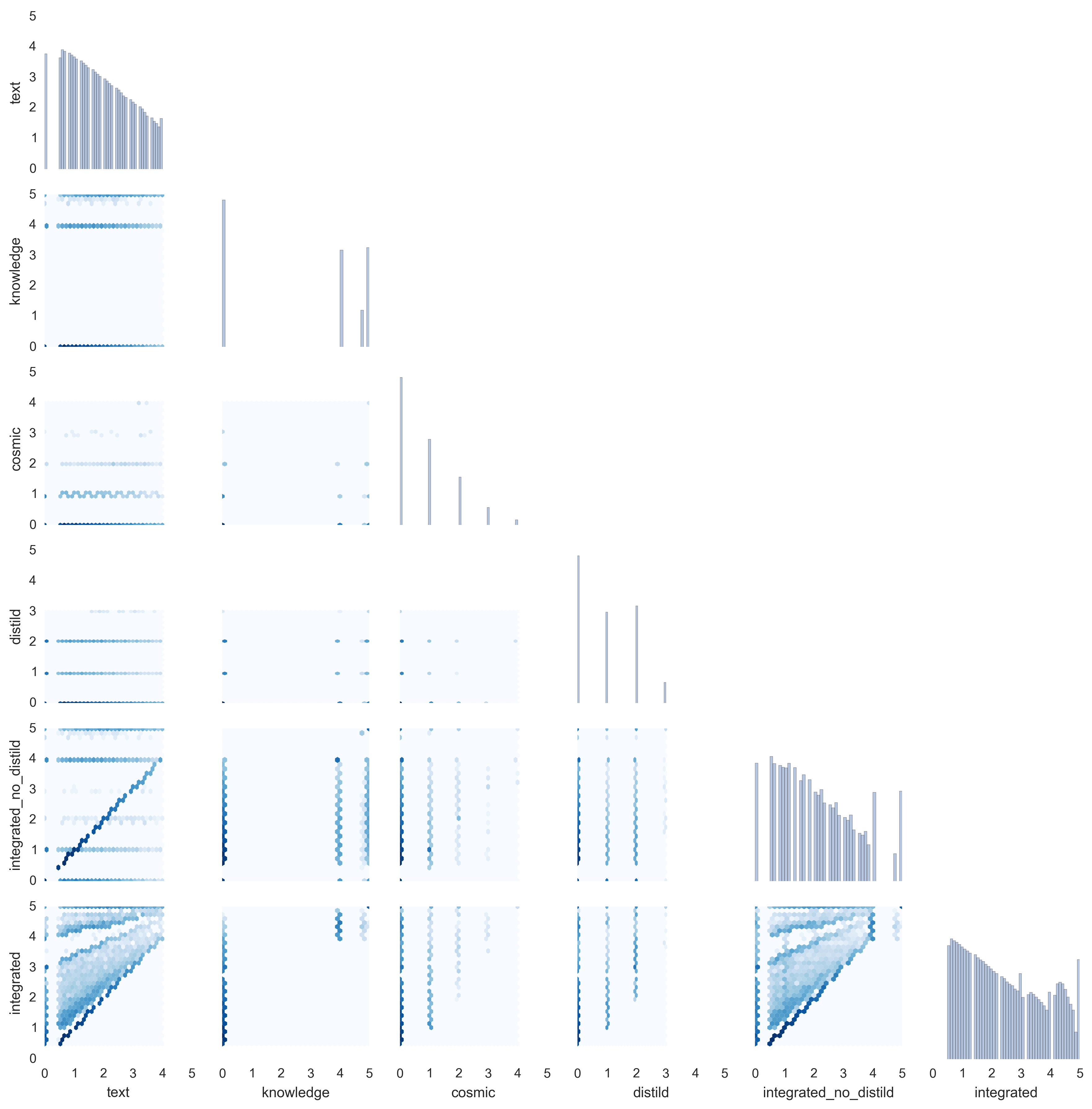
|
|
|
Status:
Completed
Views
84
Topics
Referenced by
Cite this as
Daniel Himmelstein, Lars Juhl Jensen (2015) Processing the DISEASES resource for disease–gene relationships. Thinklab. doi:10.15363/thinklab.d106
License
Share
|