|
|
|
Views
95
Topics
Referenced by
Cite this as
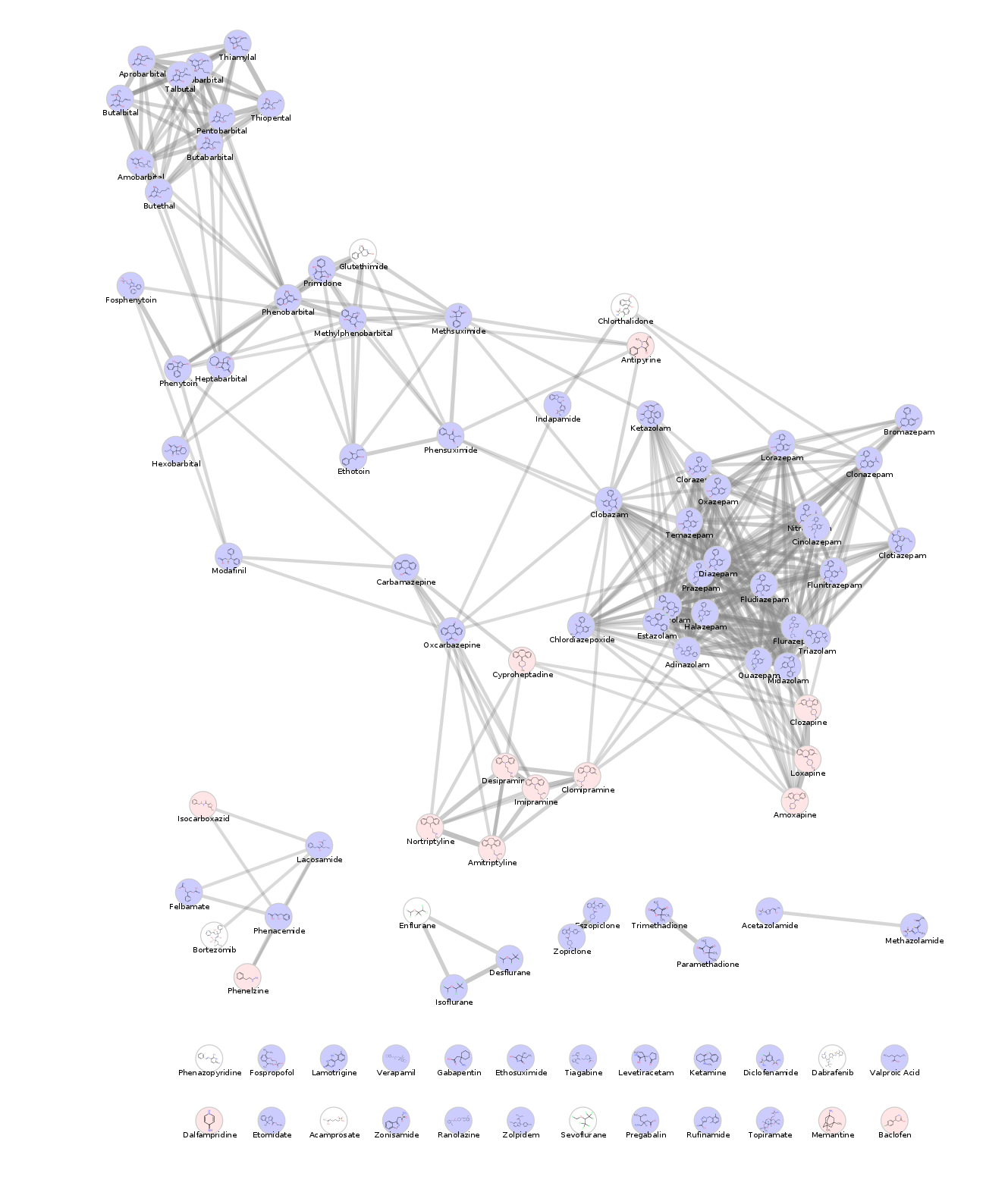
Daniel Himmelstein, Pouya Khankhanian, Alexander Pico, Lars Juhl Jensen, Scooter Morris (2017) Visualizing the top epilepsy predictions in Cytoscape. Thinklab. doi:10.15363/thinklab.d230
License
Share
|